AQUA-DUCT: Analysis of Molecular Dynamics Simulations of Macromolecules with the use of Molecular Probes [Article v1.0]
DOI:
https://doi.org/10.33011/livecoms.2.1.21383Keywords:
molecular probes, drug design, molecular dynamics, tutorialsAbstract
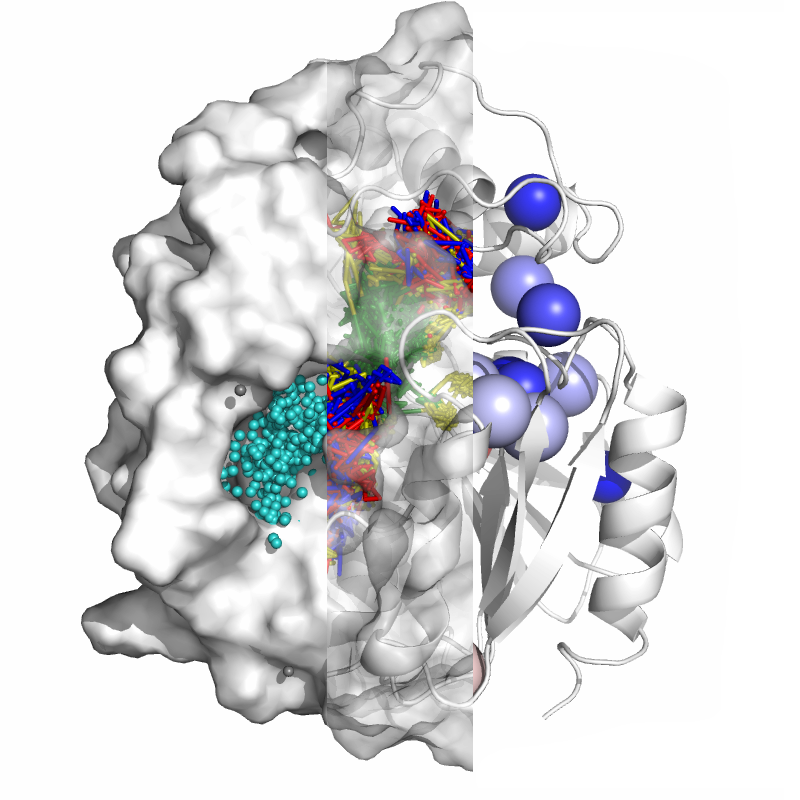
AQUA-DUCT software reverses the standard approach of the molecular dynamics simulations analysis of macromolecules, focusing on solvent, cosolvent and small ligands analysis considered as specific molecular probes instead of analysis of macromolecules atoms movement. In this article we present six basic tutorials instructing the users in the best practices for preparing, carrying out, and analysing AQUA-DUCT results in various applications. Users are expected to already have significant experience with running standard molecular dynamics simulations in any dedicated software (e.g., Amber, GROMACS, NAMD), and usage of PyMOL visualising software. The tutorials range from a basic analysis of multiple solvents trajectory used for identification of the entries/exits to the protein core, to a complex one, like identification of the key hot-spots in cosolvent MD simulations that can be used as an insight for macromolecules description, analysis, re-engineering, and for drug design.